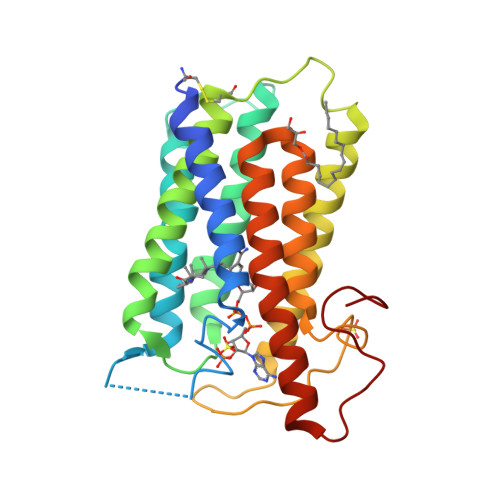
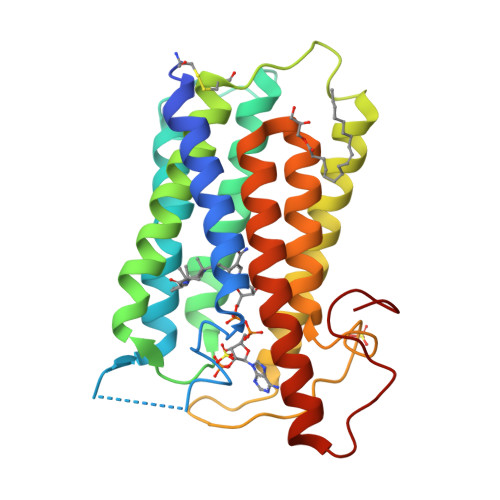
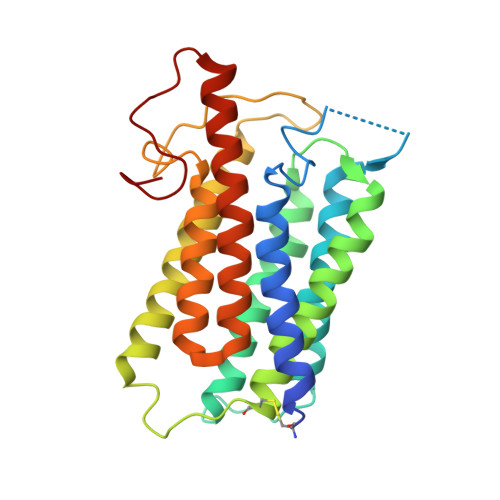
Structure of human steroid 5 alpha-reductase 2 with anti-androgen drug finasteride.
Xiao, Q., Wang, L., Supekar, S., Shen, T., Liu, H., Ye, F., Huang, J., Fan, H., Wei, Z., Zhang, C.(2020) Res Sq
- PubMed: 32702725
- DOI: https://doi.org/10.21203/rs.3.rs-40159/v1
- Primary Citation of Related Structures:
7BW1 - PubMed Abstract:
Human steroid 5α-reductase 2 (SRD5α2) as a critical integral membrane enzyme in steroid metabolism catalyzes testosterone to dihydrotestosterone. Mutations on its gene have been linked to 5α-reductase deficiency and prostate cancer. Finasteride and dutasteride as SRD5α2 inhibitors are widely used anti-androgen drugs for benign prostate hyperplasia, which have recently been indicated in the treatment of COVID-19. The molecular mechanisms underlying enzyme catalysis and inhibition remained elusive for SRD5α2 and other eukaryotic integral membrane steroid reductases due to a lack of structural information. Here, we report a crystal structure of human SRD5α2 at 2.8 Å revealing a unique 7-TM structural topology and an intermediate adduct of finasteride and NADPH as NADP-dihydrofinasteride in a largely enclosed binding cavity inside the membrane. Structural analysis together with computational and mutagenesis studies reveals molecular mechanisms for the 5α-reduction of testosterone and the finasteride inhibition involving residues E57 and Y91. Molecular dynamics simulation results indicate high conformational dynamics of the cytosolic region regulating the NADPH/NADP + exchange. Mapping disease-causing mutations of SRD5α2 to our structure suggests molecular mechanisms for their pathological effects. Our results offer critical structural insights into the function of integral membrane steroid reductases and will facilitate drug development.
Organizational Affiliation:
Department of Biology, Southern University of Science and Technology, Shenzhen, Guangdong 518055, China.